RNA Disease
RNA molecules and their processing regulations are essentially involved in gene expression process.Once transcribed from genomic DNA, pre-mRNAs undergo RNA processing, including 5′-capping, polyadenylation, and RNA splicing, to become mature mRNAs, which are then transported to ribosome for translation. In addition, numerous functional RNA species, such as non-coding RNAs, have been discovered by a recent progress in RNA sequencing. On the other hand of the physiological importance of RNA molecules, their functional impairment is also associated with manifestation of various diseases. Those diseases caused by an impaired RNA function or processing are called “RNA disease”.
1. RNA splicing regulation and diseases
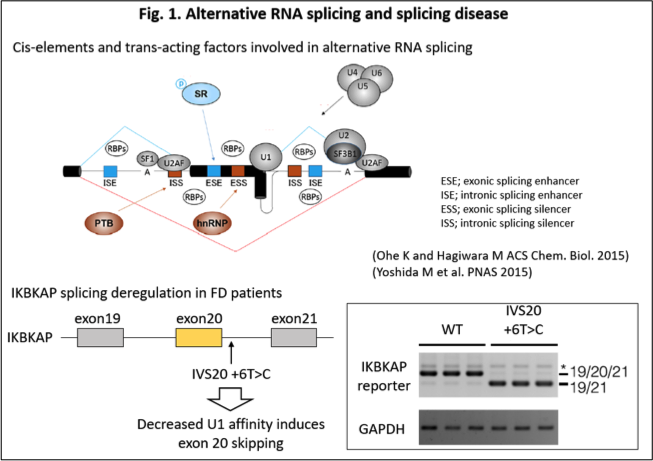
Through RNA splicing, exons are connected from pre-mRNA, while intronic sequences are removed. Most exon/introns constantly undergo splicing regulation, while some are regulated by a specific trans-acting factor, exhibiting a cell types and/or condition-dependent regulation. The former is called constitutive RNA splicing, while the latter is called alternative RNA splicing. By far, about 30% of genetic diseases are estimated to be caused by RNA missplicing due to mutations in cis-regulatory elements and/or trans-acting factors. For example, Familial Dysautonomia (FD) is caused by the exon 20 skipping of IKBKAP gene, by the T>C mutation at 6th base of intron 20 (IVS20+6T>C), which suppresses recognition of the exon 20 donor site by U1 snRNP. The exon 20 skipping then results in reduced production of IKAP protein, causing impaired tRNA modification (Fig. 1). In addition to mechanistic study of those missplicing-associated diseases, our study is also extended to therapeutics of missplicing-associated diseases by small molecule compounds.
2. Therapeutic strategy of RNA splicing diseases by small molecule compounds
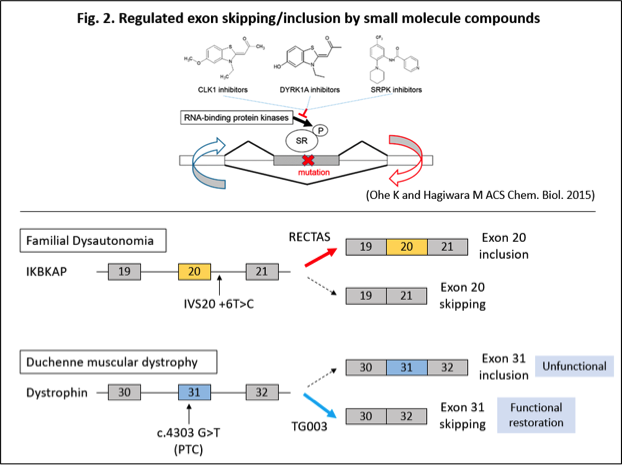
In order to manipulate disease-associated splicing regulations, we target control of serine/arginine-rich splicing factors (SR protein) activities. SR proteins are trans-acting splicing factors, known to regulate alternative RNA splicing, and their activity is regulated by phosphorylation of RS domain by SR protein kinases (SRPKs) and cdc-like kinases (CLKs) to promote a spliceosomal recognition. Our laboratory have developed small molecule inhibitors targeting those kinases, such as SRPIN340 for SRPKs (Fukuhara T et al. PNAS 2006), TG003 and related analogues for CLKs (Muraki M et al. J. Biol. Chem. 2004; Ogawa Y et al. Nat. Commun. 2010, Boisson B et al. J. Clin. Invest. 2018) and demonstrated their therapeutic effect on splicing diseases by manipulating SR protein activities. For instance, in Duchenne muscular dystrophy caused by a c.4303G>T mutation, which creates premature stop codon within the exon 30, inhibition of CLK by TG003 induces skipping of the mutated exon 30 to restore in frame-deleted functional dystrophin (Fig. 2) (Nishida A et al. Nat. Commun. 2011). In addition, we have demonstrated inhibition of SRPKs by SRPIN340 leads to a suppressed viral RNA transcription, and angiogenesis in cancer cells are also compromised through VEGF splicing regulation (Dong Z et al. Mol. Vis. 2013; Amin EM et al. Cancer Cell 2011; Nowak DG et al. J. Biol. Chem. 2010; Fukuhara T et al. PNAS 2006). Moreover, we have developed RECTAS, a compound that selectively promotes IKBKAP exon 20 inclusion through chemical library screening, and confirmed therapeutic effect in FD patient cells (Fig. 2) (Yoshida M et al. PNAS 2015). We have also confirmed those compounds impact only a limited population of cellular transcripts without affecting majority of normal splicing. Therefore, we believe that splice-targeting therapeutics can be achieved by applying those small molecule compounds.
3. Research on splicing-modifying compounds and target diseases using SPREADD
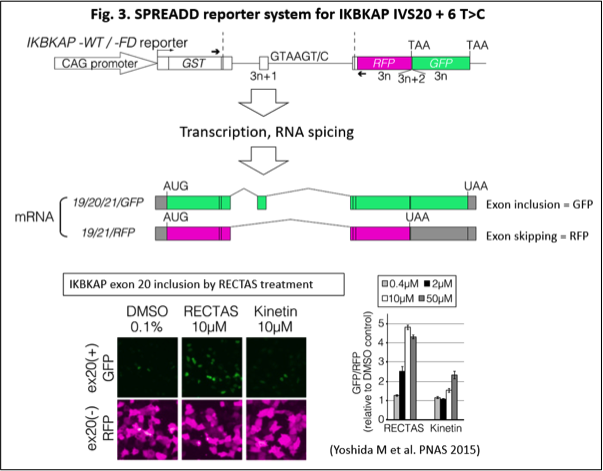
We have developed SPREADD (splicing reporter assay for disease genes with dual color) reporter system for an efficient screening of splice-regulating compounds (Fig. 3) (Yoshida M et al. PNAS 2015; Takeuchi A et al. PLOS ONE 2010; Kuroyanagi H et al. Nat. Protocol 2010). In this system, an inclusion status of a specific exon is monitored by scanning fluorescent intensities of GFP and RFP, which are switched for expression due to a frameshift. We are applying SPREADD system for a wide range of disease-associated splicing events to evaluate splice-regulating compounds and functional studies. We also investigate splice-related disease model for a therapeutic evaluation, using iPS cells and genome editing technology by CRISPR/Cas9. Through these efforts, we try to provide therapeutic compounds for splicing-associated diseases in the near future.